HBAT 2 (Hydrogen Bond Analysis Tool 2)#
A Python package to automate the analysis of potential hydrogen bonds and similar type of weak interactions in macromolecular structures, available in Protein Data Bank (PDB) file format. HBAT uses a geometric approach to identify molecular interactions by analyzing distance and angular criteria.
Supported Interaction Types:
Hydrogen Bonds: Classical
N-H···O,O-H···O, and weakC-H···OinteractionsHalogen Bonds:
C-X···Ainteractions (X = Cl, Br, I)π Interactions:
X-H...πandC-X···πinteractions with aromatic rings (Phe,Tyr,Trp,His, etc.)π-π Stacking: Aromatic ring-ring interactions (parallel, T-shaped, offset)
Carbonyl Interactions:
n→π*interactions between carbonyl groupsn-π Interactions: Lone pair interactions with aromatic
πsystems
Announcement
HBAT 2 Web Interface is Live! Try it out at hbat-web.abhishek-tiwari.com













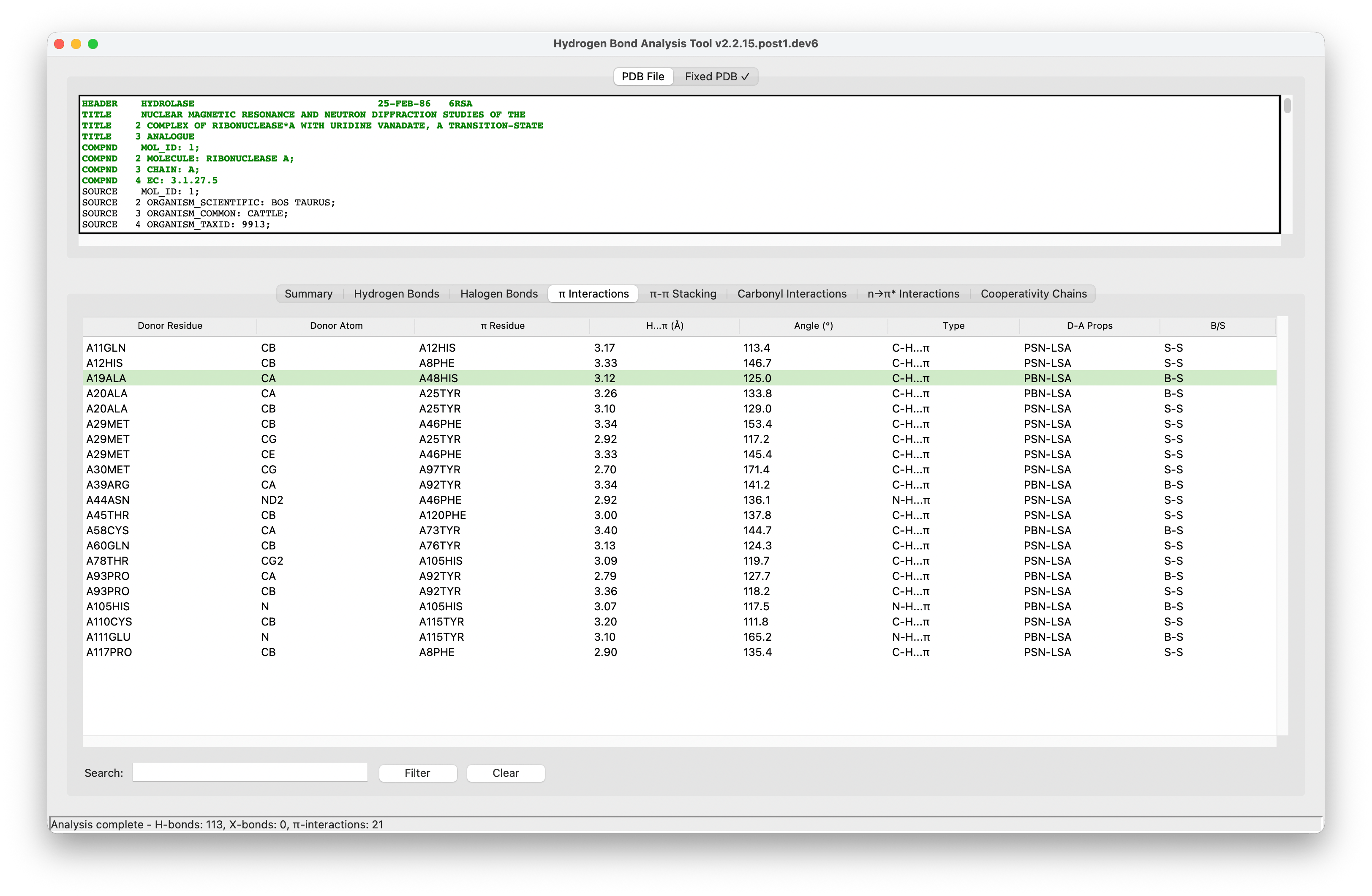
HBAT Desktop (Mac, Windows, Linux).#
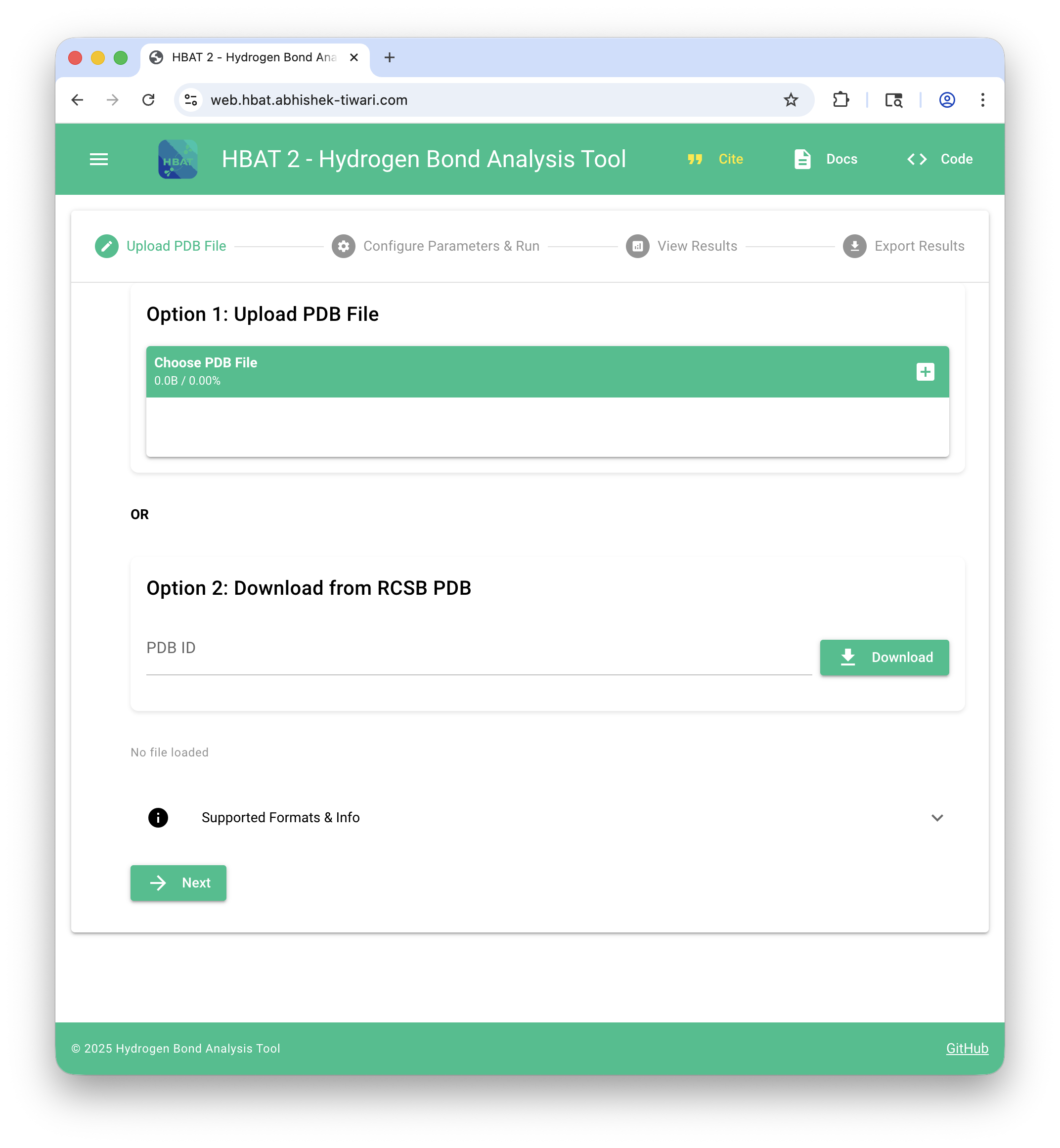
HBAT Web (https://hbat-web.abhishek-tiwari.com)#
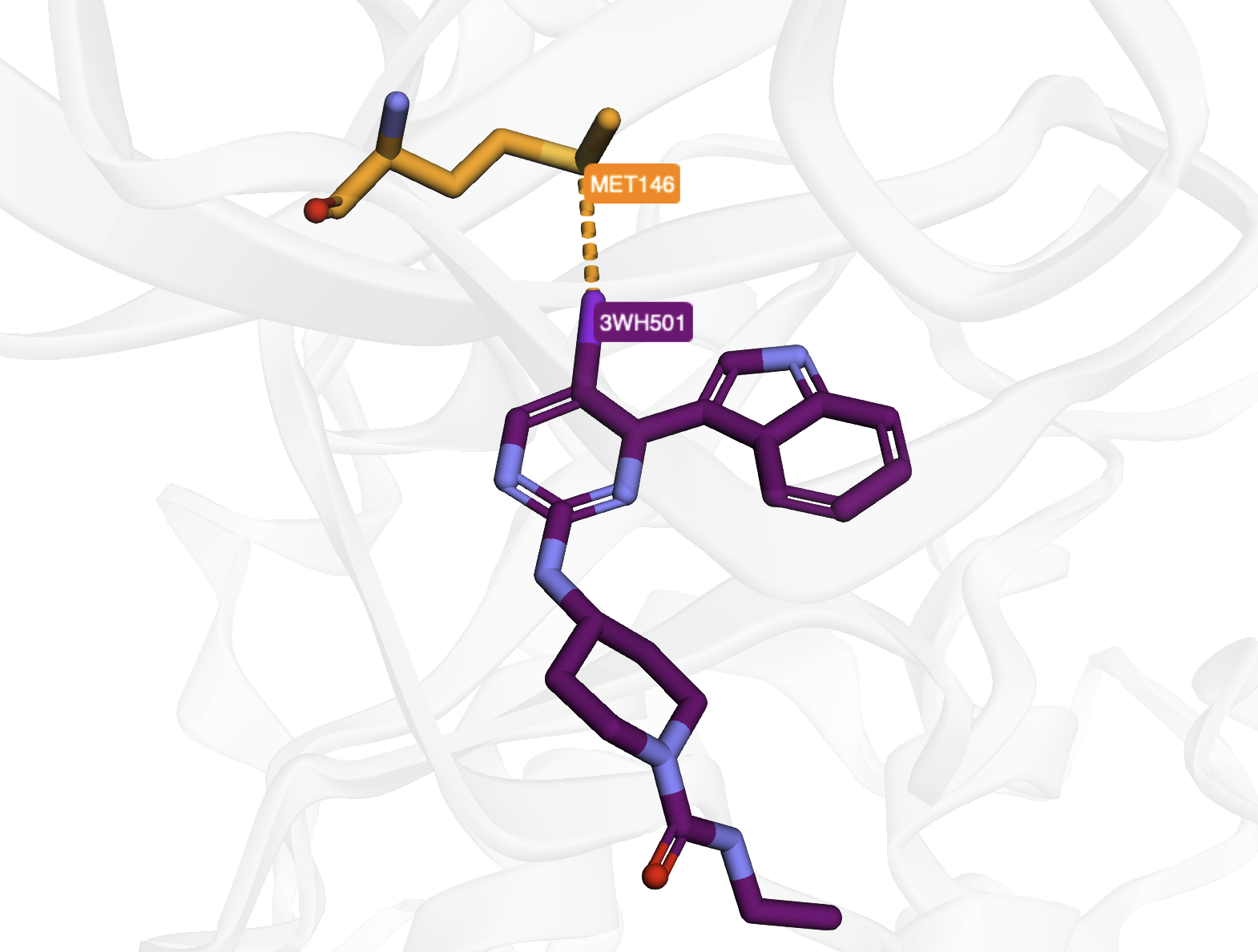
Visualizing interactions using HBAT Web (Halogen Bond in PDB Entry 4x21).#
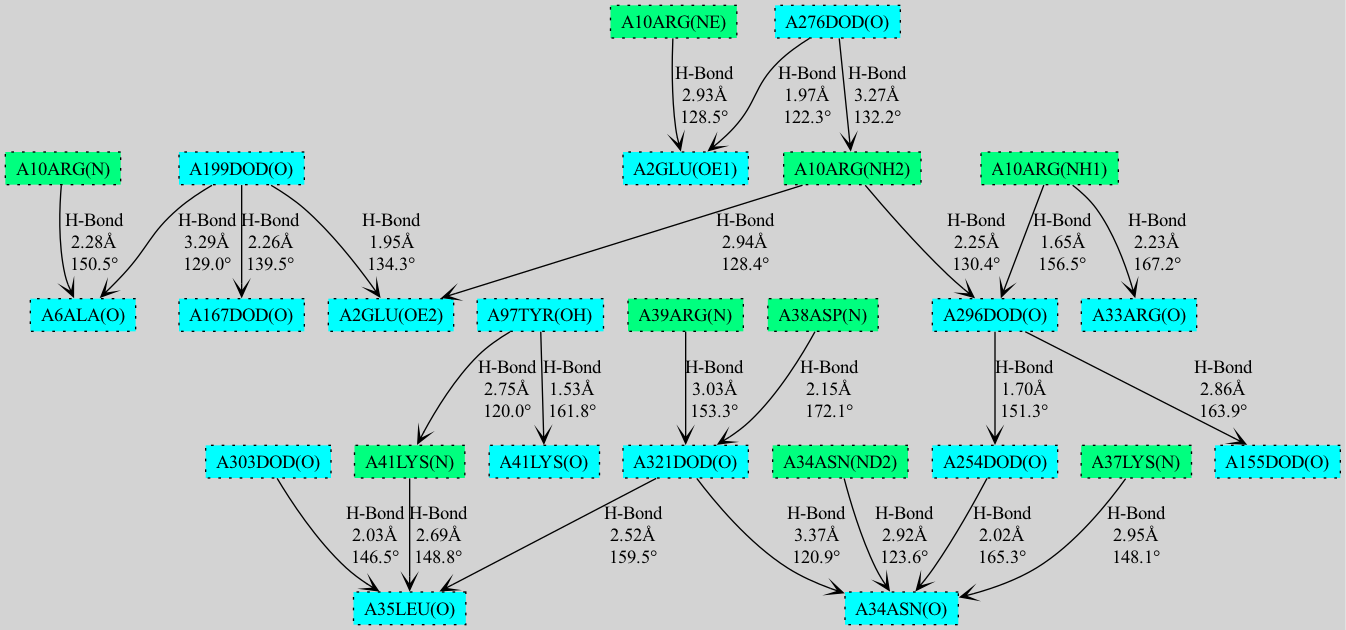
Cooperativity chain detection and visualization (PDB Entry 6RSA).#
Background#
HBAT 2 is a modern Python re-implementation of the original Perl-based tool developed by Abhishek Tiwari and Sunil Kumar Panigrahi. HBAT v1 can still be downloaded from SourceForge however Perl version is not maintained anymore.
Highlights of HBAT 2#
Detect and analyze potential hydrogen bonds, halogen bonds, π interactions, π-π stacking, carbonyl interactions, and n-π interactions
Automated PDB fixing with OpenBabel and PDBFixer integration
Support graphical (tkinter), command-line, and programming API interfaces
Use graphical interfaces for interactive analysis, CLI/API for batch processing and automation
Cooperativity chain visualization using NetworkX/matplotlib and GraphViz
Export cooperativity chain visualizations to PNG, SVG, PDF formats
Built-in presets for different structure types (high-resolution, NMR, membrane proteins, etc.)
Customizable distance cutoffs, angle thresholds, and analysis modes.
Multiple Output Formats: Text, CSV, and JSON export options
Optimized algorithms for efficient analysis of large structures
Cross-Platform: Works on Windows, macOS, and Linux.
Cite HBAT & HBAT 2#
@article{tiwari2007hbat,
author = {Tiwari, Abhishek and Panigrahi, Sunil Kumar},
doi = {10.3233/ISI-2007-00337},
journal = {In Silico Biology},
month = dec,
number = {6},
title = {{HBAT: A Complete Package for Analysing Strong and Weak Hydrogen Bonds in Macromolecular Crystal Structures}},
volume = {7},
year = {2007}
}
@misc{tiwari_2025_17645321,
author = {Tiwari, Abhishek},
title = {HBAT 2: A Python Package to analyse Hydrogen Bonds and Other Non-covalent Interactions in Macromolecular Structures},
month = nov,
year = 2025,
publisher = {Zenodo},
doi = {10.5281/zenodo.17645321},
url = {https://doi.org/10.5281/zenodo.17645321},
}
Tiwari, A., & Panigrahi, S. K. (2007). HBAT: A Complete Package for Analysing Strong and Weak Hydrogen Bonds in Macromolecular Crystal Structures. In Silico Biology, 7(6). https://doi.org/10.3233/ISI-2007-00337
Tiwari, A. (2025). HBAT 2: A Python Package to analyse Hydrogen Bonds and Other Non-covalent Interactions in Macromolecular Structures. Zenodo. https://doi.org/10.5281/zenodo.17645321
User Guide